Thoughts
- RAG & reading paper (done in scChat)
- Based on paper, presume possible clusters and markers. Run initial cluster based only on marker genes to establish a rough cell type assignment. Then refine clustering with the full gene set while maintaining the initial structure
- Use distances between clusters (e.g. UMAP)?
- Use gene expr proportion in clusters? But consider drop out
- Multiple AI average (scTriangulate, leverages cooperative game theory (Shapley Value))
- Generate report
Questions
- How to deal with multiple results from AIs
- Drop out
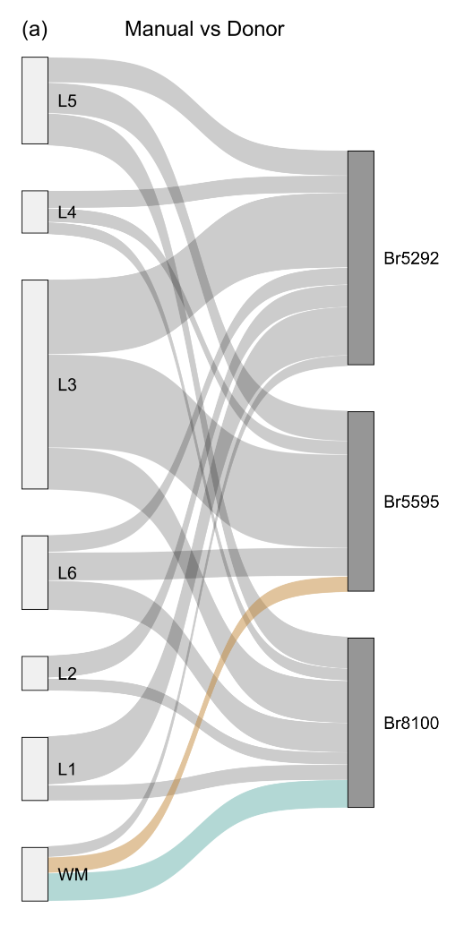
Evaluation
- Sankey plot. See A web app for comparing cell annotations or clusterings • tagtango